A new tool to identify linear polyubiquitin-modified proteins
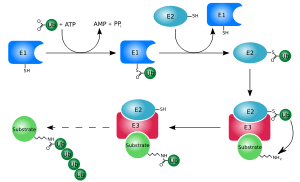
Ubiquitination is a Post-Translational Modification (PTM) that modifies proteins with Ubiquitin (Ub). The discovery that ubiquitin chains target proteins to the proteasome, which degrades and recycles proteins, was honored with the Nobel Prize in chemistry in 2004. Indeed, polyubiquitination regulates lots of cellular processes, mainly protein stability, inflammation, DNA repair and immune response. Ubiquitination is carried out in three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3), respectively. The result of this sequential cascade mainly binds ubiquitin to lysine residues on the protein substrate via an isopeptide bond, but also cysteine residues through a thioester bond, serine and threonine residues through an ester bond, or the amino group of the protein’s N-terminus via a peptide bond. Differently linked chains have specific effects on the protein to which they are attached, caused by differences in the conformations of the protein chains. K29-, K33-, K63- and M1-linked chains have a fairly linear conformation, and they are known as open-conformation chains. K6-, K11-, and K48-linked chains form closed conformations.
Ubiquitin-Binding Domains
Some proteins can specifically bind to ubiquitin via ubiquitin-binding domains (UBD). The distances between individual ubiquitin units in chains differ between lysine 63- and 48-linked chains. The UBDs exploit this by having small spacers between ubiquitin-interacting motifs that bind lysine 48-linked chains (compact ubiquitin chains) and larger spacers for lysine 63-linked chains. The machinery involved in recognising polyubiquitin chains can also differentiate between K63-linked chains and M1-linked chains, demonstrated by the fact that the latter can induce proteasomal degradation of the substrate.
Inhibits activity of E3 ligase
Furthermore, identification of E3 ligase substrates is critical to understand its implication in human diseases since deregulation of E3-substrate interactions are often served as major cause in many. Finding a specific molecule that selectively inhibits the activity of a certain E3 ligase and/or the protein-protein interactions implicated in the disease remains as one of the important and expanding research area. Moreover, as ubiquitination is a multi-step process with various players and intermediate forms, consideration of the much complex interactions between components needs to be taken heavily into account while designing the small molecule inhibitors.
Identify linear polyubiquitin-modified proteins
Kliza et al. recently published in Nature Methods a new method to identify linear polyubiquitin-modified proteins (Kliza et al. Nat Methods. 2017 May;14(5):504-512.). Indeed, current methods for studying lysine-based polyubiquitination are not suitable for the detection of linear polyubiquitin-modified proteins. Kliza et al. describe an approach for the specific enrichment of linear poly-Ubiquitin-modified substrates, by using a internally tagged Ubiquitin. They chose the STREP-tag II because this short, almost biochemically inert tag strongly interacts with Streptavidin Strep-Tactin under denaturating conditions. Thus, this technique using a K-less internally tagged Ubiquitin pool allows to identify linear polyubiquitinated targets in a simple and robust manner. This new tool should contribute to the understanding of the largely underestimated spectrum of linear polyubiquitination, expanding the current view of its cellular roles.